基因交互作用大大影響胰臟癌病人對藥物的反應
胰臟癌是非常惡性的腫瘤,病人的五年存活率在8-10%左右。因為胰臟癌不易早期發現,只有20%左右的病人可以開刀治療,在轉移的病人,目前化療大多是以Gemcitabine組合的藥物當作第一線的治療,然而藥物的反應率低,所以胰臟癌病人的存活率在近20年並無明顯的改善。
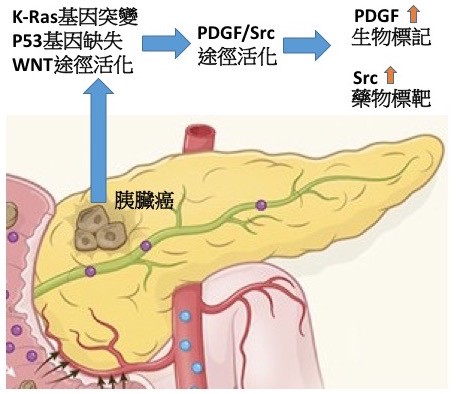
精準醫學是癌症治療上非常重要的一環,不同的胰臟癌組織存在不同的基因缺陷,這些基因異常可單獨或協同活化不同的訊息傳遞途徑,所以癌症病人對化學或標靶治療的反應性有很大的差別。如何藉由細胞實驗,動物模式及病人基因檢測找到藥物反應的生物標記(Biomarker)是基礎研究人員與臨床醫師的共同目標。在胰臟癌中,變異最高的兩個基因是K-ras與p53。然而除了這兩個基因外,常有第三個基因缺陷,本校洪文俊副校長領導的團隊發現WNT/β-catenin途徑的變異存在約20%的胰臟癌病人。團隊建立了基因轉殖鼠的動物模式,研究發現相較於只有K-ras與p53變異的基因轉殖鼠,具有第三個WNT/β-catenin途徑基因缺陷的老鼠會特異性的增加血小板衍生生長因子(PDGF)基因轉錄,進而產生大量的PDGF來刺激胰臟癌細胞與星狀細胞生長,快速促進胰臟癌惡化轉移。進一步,團隊與國家衛生研究院及國立成功大學醫學院合作,在胰臟癌病人的血液與腫瘤組織印證了臨床相關性。更重要的,藉由分子機制探討及藥物篩選,團隊發現PDGF途徑特異性活化下游的訊息傳遞分子Src,所以具有三個基因缺陷的老鼠對Src抑制劑特別敏感,顯示此類藥物對具有特定基因變異的胰臟癌病人可能有更顯著的治療效果。此外血液PDGF的含量也可以是當作篩選病人的生物標記。本研究建立了新穎的胰臟癌動物模式來探討變異基因之交互作用對胰臟癌惡化與轉移的影響,同時找到了一個血液生物標記可用於篩選適合於標靶治療的胰臟癌病人(如圖示),達到精準醫療的目的,相關研究成果發表於國際期刊Theranostics。
本篇為高雄醫學大學2019年傑出論文及2019年月傑出論文1月份得獎文章,代表作者為醫學院醫學研究所洪文俊教授。
基因交互作用大大影響胰臟癌病人對藥物的反應
胰臟癌是非常惡性的腫瘤,病人的五年存活率在8-10%左右。因為胰臟癌不易早期發現,只有20%左右的病人可以開刀治療,在轉移的病人,目前化療大多是以Gemcitabine組合的藥物當作第一線的治療,然而藥物的反應率低,所以胰臟癌病人的存活率在近20年並無明顯的改善。
精準醫學是癌症治療上非常重要的一環,不同的胰臟癌組織存在不同的基因缺陷,這些基因異常可單獨或協同活化不同的訊息傳遞途徑,所以癌症病人對化學或標靶治療的反應性有很大的差別。如何藉由細胞實驗,動物模式及病人基因檢測找到藥物反應的生物標記(Biomarker)是基礎研究人員與臨床醫師的共同目標。在胰臟癌中,變異最高的兩個基因是K-ras與p53。然而除了這兩個基因外,常有第三個基因缺陷,本校洪文俊副校長領導的團隊發現WNT/β-catenin途徑的變異存在約20%的胰臟癌病人。團隊建立了基因轉殖鼠的動物模式,研究發現相較於只有K-ras與p53變異的基因轉殖鼠,具有第三個WNT/β-catenin途徑基因缺陷的老鼠會特異性的增加血小板衍生生長因子(PDGF)基因轉錄,進而產生大量的PDGF來刺激胰臟癌細胞與星狀細胞生長,快速促進胰臟癌惡化轉移。進一步,團隊與國家衛生研究院及國立成功大學醫學院合作,在胰臟癌病人的血液與腫瘤組織印證了臨床相關性。更重要的,藉由分子機制探討及藥物篩選,團隊發現PDGF途徑特異性活化下游的訊息傳遞分子Src,所以具有三個基因缺陷的老鼠對Src抑制劑特別敏感,顯示此類藥物對具有特定基因變異的胰臟癌病人可能有更顯著的治療效果。此外血液PDGF的含量也可以是當作篩選病人的生物標記。本研究建立了新穎的胰臟癌動物模式來探討變異基因之交互作用對胰臟癌惡化與轉移的影響,同時找到了一個血液生物標記可用於篩選適合於標靶治療的胰臟癌病人(如圖示),達到精準醫療的目的,相關研究成果發表於國際期刊Theranostics。

本篇為高雄醫學大學2019年傑出論文及2019年月傑出論文1月份得獎文章,代表作者為醫學院醫學研究所洪文俊教授。

